
On-silica methordary for protein domain modification
Background
We are at the forefront of developing next-generation downstream processing technologies, specializes in innovative matrices and microspheres commonly used for purifying target macromolecules through specific affinity binding, typically derived from specific protein-protein interactions or enzymatic activities.
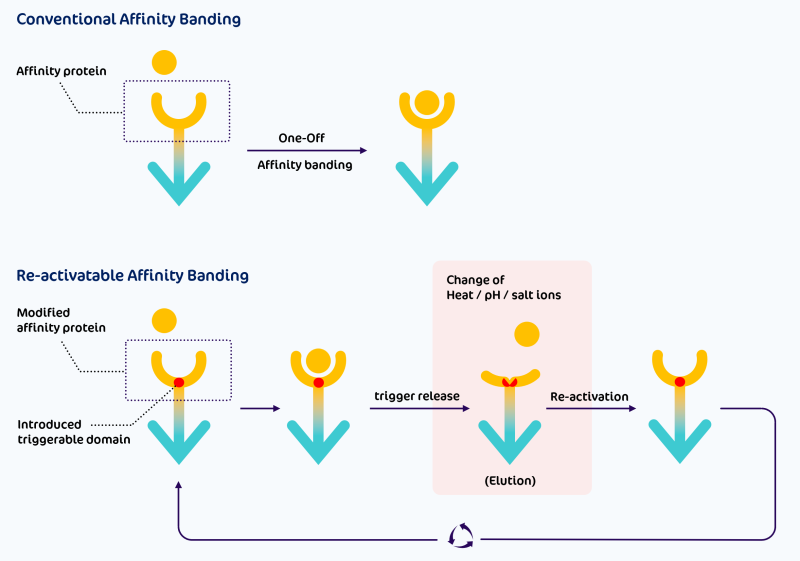
To meet the rapidly evolving demands of advanced medical devices and industrial downstream processing, reusable (elutionable) approaches are emerging as the trend for next-generation matrices and microspheres. In contrast, conventional single-use (irreversible binding) affinity protein/ligand systems are becoming less favourable due to the uneconomical waste of finely-produced matrices and precious microspheres.
For example, traditionally, VHH antibodies irreversibly bind to target substrates (antigens). When these antibodies are cross-linked onto matrices and incorporated into medical devices such as hemoperfusion systems, these devices are typically disposable and often do not achieve optimal binding capacity and longevity. However, if VHH antibodies are structurally modified to allow for temperature/pH/salt-triggered release mechanisms, the fully bound and capacity-exhausted core matrices and microspheres of these medical devices can be reactivated using warm, acid-basic, or salty buffer elution, enabling the entire device to operate in a long-term repeatable or even continuous mode, which is economically advantageous and welcomed.
This innovation is also welcomed in other sectors' downstream processing.
Directions and Goals
One of our directions is to develop a toolkit that helps achieve modifications to the existing functional proteins in our arsenal. It can be universally applied to our proteins of interest, as well as those of our peers, to achieve complex functions and finely timed and space-controlled treatments or catalysis.
The toolkit is a software that uses AlphaGo for its output calibration and returns next-round calculations to predict and simulate any possible derivative conformation of protein activity sites and introduce desired functions. The initial raw idea is the Monte Carlo method, assisted by neural network learning. Using the types and coordinates of key amino acids in the known functional domains of the target protein as boundary conditions, the software creates various structures by relinking key amino acids into a sequence that allows easy expression while maintaining the activity of the original functional domains and introducing desired activity conformation. The candidate structures with the lowest internal energy (most stable) are then selected.
Use and impact
This software will help us optimize and modify the conformation of the target enzyme, affinity protein, or antibody's key activity domain to enhance specific enzymatic or affinity performance. Additionally, the software will have the capability to adjust the essential domain's conformation to achieve pH, ionic strength, salt, or temperature-triggered binding and release mechanisms by modifying amino acid residues, introducing solvent-sensitive amino acids, or incorporating pairs of amino acid residues, such as a disulfide bond between Cys-Cys, to create a triggerable conformation. This approach may evolve to realize triggerable enzymatic activities and conformation deformation-driven specific affinity for more advanced applications. Triggerable specific affinity binding and release will make the affinity matrix reusable, high-throughput, and continuously operational, significantly boosting the efficiency and convenience of downstream processes. It will also enable one-shot binding and differentiated release by elution for various target molecules.
This work is critical for the development of next-generation hemoperfusion medical devices, external organs, and complex cellular therapeutic devices. It will also serve as a foundational infrastructure for future developments in protein science and macromolecule expression.
Collaborative Research
Neopuriph Pty Ltd conducts the majority of its R&D activities within the Biomanufacturing Research Group at the University of Adelaide. These efforts are primarily carried out by PhD students and postdocs, who are either fully or jointly funded by the company and the university. We aim to expand this model to include collaboration with scholars and research groups worldwide, fulfilling the above-mentioned research and tasks, and further advancing our innovative work in downstream processing technologies.